Molecular Biology 2019
1.) Answer any ten questions from the following:
a) Define stop codon.
Ans→ A stop codon is a sequence of three nucleotides in mRNA that provide a signal to terminate the protein synthesis.
UAG, UAA and UGA are the stop codons.
b) What is microRNA?
Ans→ MicroRNA (miRNA) are small, highly conserved non-coding RNA molecules involved in the regulation of gene expression.
c) State the function of DNA Gyrase?
Ans→ DNA Gyrase helps in unwinding of DNA by by removing the strain during the replication process.
d) State the function of anticodon loop at t-RNA.
Ans→ Anticodon loop of the t-RNA helps is recognition of the mRNA codon leading to the formation of peptide bonds between the amino acids.
e) What is the significance of Northern blotting?
Ans→ (i) RNA Detection: Identifies and measures specific RNA molecules in a sample.
(ii) Gene Expression Analysis: Helps determine gene expression levels across different conditions or tissues.
f) What is ribo switch?
Ans→ Riboswitches are structured non-coding RNA domains that selectively bind metabolites and control gene expression.
g) Write the Wobble hypothesis?
Ans→ Wobble hypothesis explains that the third base pairing varies in respect of codon like ‘G’ may pair with ‘C’ or ‘U’ where one amino acid can be coded by more than one codon.
h) State the function of DNA Pol-I
Ans→ DNA Pol-I involved in DNA repair mechanism and the removal of RNA primers during DNA replication.
ⅰ) What is conservative replication concept?
Ans→ In conservative replication concept, the DNA replicates and produces two DNA copies where one is entirely new DNA and the other is made of old DNA.
j) Mention the role of sigma (o) factor is prokaryotes.
Ans→ Sigma factor is a Key component that associates with the RNA polymerase core enzyme to form RNA polymerase holoenzyme which is required for initiation of transcripts.
k) State the importance of primer in PCR.
Ans→ Primers provide the 3′-OH group at the end from where DNA Pol can add the nucleotides and form the new strand.
I) What is polyadenylation?
Ans→ Polyadenylation is the addition of poly/multiple adenine bases at the ‘3’ end of RNA transcript (mRNA) by Poly A polymerase (PAP) enzyme.
m) How many nucleotides.
Ans→ No nucleotides are being added by ligase enzymes. It can only form phosphodiester bonds between the adjacent nucleotides in DNA duplex.
n) what is intron lariat?
Ans→ An intron lariat is a loop-like structure formed during RNA splicing from pre-mRNA.
o) What is palindromic sequence? Give example.
Ans→ Palindromic sequence are referred as the sequence of nucleotides in the DNA where the sequence of one strand is the same as the reverse complementary sequence.
Example: ACCTAGGT is a palindrome. The complementary strand have TGGATCCA.
2.) Answer any five of the following questions:
a) What is the role of EF-Ts and EF-Tu factor during protein synthesis?
Ans→
b) What is Klenow fragment? Which enzyme is known as Kornberg enzyme?
Ans→ Klenow fragment is the large proteolytic fragment of DNA pol I of E. coli which retains polymerization and 3’→5′ exonuclease activity, but has lost 5’→3’ exonuclease activity.
DNA polymerase I is known as Kornberg enzyme.
c) Differentiate spliceosome and replisome.
Ans→
| Spliceosome | Replisome | |
| 1) Function | Involved in RNA splicing | Involved in DNA replication |
| 2) Components | Composed of (snRNAs) and associated proteins (forming snRNPs). | Consists of enzymes like DNA polymerase, helicase, primase, and other proteins. |
| 3) Location of Activity | Works in the nucleus of eukaryotic cells. | Works at the replication fork in both prokaryotic and eukaryotic cells. |
| 4) Process | Cuts and rejoins RNA sequences to remove non-coding introns. | Unwinds DNA and synthesizes new strands complementary to the original DNA templates. |
d)Briefly describe the Sanger DNA sequencing method.
Ans→ Sanger sequencing is a DNA sequencing method that uses dideoxynucleotide chain terminators to halt DNA synthesis at specific bases. By labeling these terminators with fluorescent dyes and running the resulting DNA fragments through gel or capillary electrophoresis, the DNA sequence can be read based on the order of fragment sizes.
e) What do you mean by closed & open promoter complex in transcription?
Ans→ In transcription, a closed promoter complex is when RNA polymerase binds to the promoter without unwinding the DNA. An open promoter complex forms when the DNA unwinds at the promoter region, allowing RNA polymerase to begin RNA synthesis.
f) what is 5′ capping? State the significance.
Ans→ 5’ capping is a process of addition of 7 methyl guanosine cap at the 5 end of RNA transcript (mRNA) by help of RNA (guanine-7)-methyl transferase.
Significance: 5′ cap protects the m newly synthesised mRNA from degradation, provides stability and helps in recognition by ribosomes.
g) Briefly describe the attenuation process.
Ans→ Attenuation process involves a provisional stop signal (attenuator), by some bacterial operons that results in premature termination of transcription.
Stop signal that prevents the proper function of ribosomal complex leading termination.
h) What are the different sites of ribosomes? Give their function.
Ans→ Aminoacyl site (A-site), Peptidyl site (P-site) and Exit Site (E-site) are the different sites of ribosome.
Functions:
- A-site: A-site accepts an incoming tRNA bound to an amino acid.
- P-site: P-site holds a tRNA that makes a growing polypeptide.
- E-site: E site provides the exit path to tRNA from ribosomes.
Molecular Biology 2020
a) What is denaturation?
Ans→ Denaturation is the process where proteins or nucleic acids lose their natural structure due to external stress, like heat or chemicals, disrupting their function.
b) What is Central dogma?
Ans→ Central dogma is a theory stating that genetic Information flows only in o one direction, from DNA to RNA to protein, by DNA replication, transcription & translation process.
c) What is ‘Replication bubble’?
Ans→ A replication bubble is a region in the DNA molecule where the double helix unwinds and separates to allow for DNA replication to occur.
d) Define Nucleotide excision repair?
Ans→ Nucleotide excision repair is a DNA repair mechanism that detects and removes damaged or incorrect nucleotides from the DNA strand and replaces it with the correct sequence.
e) What is ‘Pribnow box’?
Ans→ Pribnow box also known as the -10 sequence is is a short DNA sequence (TATAAT) found is the promoter region of genes in prokaryotes.
It serves as a recognition site for RNA pol during transcription initiation.
f) Define Endonuclease?
Ans→ Endonuclease is an enzyme that cleaves or cuts DNA or RNA at specific internal positions other than at the ends.
Involved in – DNA repair, restriction digestion, & RNA splicing.
g) What do you mean by C-3′ end of a polynucleotide chain?
Ans→ The C-3′ end of a polynucleotide chain is the end where the third carbon of the sugar molecule has a free hydroxyl (-OH) group, allowing it to form bonds with other nucleotides.
h) What is ‘RNA primer?
Ans→ RNA primer is a short segment of RNA that serves as a starting point for DNA replication, providing a free 3 end where DNA polymerase can synthesizing a near DNA strand.
ⅰ) Write the significance of post transcriptional modification.
Ans→(i) Helps in export of mRNA from nucleus to cytoplasm from degeneration by endonucleases.
(ii) Provides recognition site to ribosomes.
j) Difference between exon and intron.
Ans→
| Exon | Intron | |
| 1) Definition | Coding regions of a gene that are transcribed and translated into proteins. | Non-coding regions of a gene that are transcribed but removed during RNA splicing. |
| 2) Presence | Retained in mature mRNA after splicing. | Absent in mature mRNA. |
| 3) Function | Carry information for protein synthesis. | Regulate gene expression or play roles in alternative splicing |
k) Write down the full form of VNTR.
Ans→ The full form of VNTR is Variable Number of Tandem Repeats.
l) What is the function of a poly A tail?
Ans→ (i) Facilitate Stability: It helps to protect the mRNA from degradation by exonucleases.
(ii) mRNA transport: The poly A tail assists in the export of the mature mRNA from the nucleus to cytoplasm.
m) What is the significance of Western blot?
Ans→ (i) To separate and identify proteins within complex mixture.
n) By which molecular technique we can rapidly isolate and purify the genomic and plasmid DNA.
Ans→ Phenol Chloroform extraction method.
o) What is the full form of dNTP?
Ans→ dNTP- Deoxynucleotide triphosphate.
2.a) What is ‘taq-polymerase’?
Ans→ Taq polymerase is a heat-resistant DNA polymerase enzyme derived from a bacterium called Thermus aquaticus. It’s widely used in PCR to amplify DNA because it can withstand high temperatures.
b) Distinguish between prokaryotic and eukaryotic transcription.
Ans→
| Prokaryotic Transcription | Eukaryotic Transcription | |
| 1) Location | Occurs in the cytoplasm. | Occurs in the nucleus. |
| 2) RNA Polymerase | Single type of RNA polymerase. | Three types (RNA polymerase I, II, and III) for different RNA classes. |
| 3) Initiation | Requires sigma factor for promoter recognition. | Requires multiple transcription factors and promoter elements like TATA box. |
| 4) Coupling | Transcription and translation are coupled, occurring simultaneously. | Transcription and translation are separate processes. |
| 5) mRNA Processing | No post-transcriptional modifications (no capping, splicing, or polyadenylation) | Undergoes capping, splicing, and polyadenylation before becoming mature mRNA. |
c) What is telomere and its function.
Ans→ A telomere is a protective cap of repetitive DNA at the ends of chromosomes.
Its main function is to prevent chromosomes from deteriorating or fusing with each other during cell division, thereby maintaining genetic stability.
d) What is ‘operon’? Give an example.
Ans→ An Operon is a group/cluster of genes in prokaryotic organisms that are regulated, and transcribed together as a single unit.
*It includes promoter, operator and structural genes.
Example: Lac operon, Tryptophan operon
e) Mention the significance of fmet tRNA.?
Ans→ fMet-tRNA (formylmethionine tRNA) is essential for initiating protein synthesis in prokaryotes. It delivers the first amino acid, formylmethionine, to the ribosome, marking the start of translation.
f) What is the Ori C site?
Ans→ The OriC site is the specific DNA sequence in prokaryotes where replication begins, initiating the copying of the bacterial chromosome.
g) What is Polyribosome or polysome?
Ans→ A polyribosome or polysome is a cluster of ribosomes simultaneously translating a single mRNA molecule, increasing the efficiency of protein synthesis.
h) Comparison between prokaryotic and eukaryotic translation.
Ans→
| Prokaryotic Translation | Eukaryotic Translation | |
| Initiation | Initiated by fMet-tRNA binding to the small ribosomal subunit. | Initiated by Met-tRNA and scanning of the mRNA for the start codon (AUG). |
| Ribosome Size | 70S (50S + 30S subunits). | 80S (60S + 40S subunits). |
| mRNA Structure | mRNA is often polycistronic (codes for multiple proteins). | mRNA is monocistronic (codes for one protein) and has a 5′ cap and poly-A tail. |
| Translation and Transcription Coupling | Translation begins while transcription is still ongoing. | Translation occurs after transcription and mRNA processing. |
| Regulation | Simple in regulation compared with eukaryotic translation. | Complex regulation |
Molecular Biology 2021
a) What is degenerate primer?
Ans→ A degenerate primer is a mix of similar DNA sequences used in PCR to amplify genes with slight variations, ensuring binding across related sequences.
b) Which enzyme reduces the tension during the unwinding of DNA helix in front of the replication fork?
Ans→ The enzyme DNA topoisomerase (specifically DNA gyrase in prokaryotes).
c) What is TATA box?
Ans→ TATA box is a DNA sequence found in the promoter region of eucariotic genes serving as a binding site for RNA polymerase and transcription factors.
d) Define spliceosome.
Ans→ The spliceosome is a cellular machinery that removes introns (non coding region) from pre-mRNA, allowing the exons to be joined to form mRNA during splicing.
e) What is polycistronic mRNA?
Ans→ Polycistronic mRNA is a type of RNA molecule that encodes multiple genes in prokaryotes and synthesizes multiple proteins.
f) rRNA is synthesized in which portion of a cell?
Ans→ rRNA is synthesized in the nucleolus, a region within the nucleus of eukaryotic cells.
g) Write the codon for anticodon of 3′ UUA 5′.
Ans→ 5’ AAU 3’
h) In the genetic code dictionary, how many codons are used to code for all the 20 essential amino acids?
Ans→ 61 codons
i) Define chaperons.
Ans→ Chaperons on proteins that assist in the proper folding and assembly of other biomolecules, preventing misfolding and ensuring their functional integrity in cells.
j) What is rho factor?
Ans→ Rho factor is a protein in prokaryotes that helps terminate transcription by releasing the RNA transcript from the DNA template.
k) What is corepressor?
Ans→ A corepressor is a molecule that binds to a repressor protein, enabling it to inhibit gene transcription by attaching to the operator region of DNA.
l) What are the significance of CPG islands?
Ans→ CpG islands are regions of DNA rich in cytosine and guanine dinucleotides. They are important for regulating gene expression, as their methylation status can control gene activation or silencing.
m) What is holliday junction?
Ans→ holiday junction is a four way junction in DNA connecting two duplex DNA structure which is a key intermediate in homologous recombination.
n) What is SOS response?
Ans→ SOS response is the response when DNA is extensively damaged and the synthesis of a repairing mechanism.
o) What do you mean by codon degeneracy?
Ans→ Codon degeneracy refers the redundancy of genetic code where multiple codons code for the same amino acid.
- Answer any five of the following questions:
a) What is the difference between a deoxyribonucleotide and dideoxyribo- nucleotide?
Ans→
| Deoxyribonucleotide | Dideoxyribonucleotide | |
| 1) Structure | Contains a deoxyribose sugar with a hydroxyl group (-OH) at the 3′ carbon. | Contains a deoxyribose sugar with no hydroxyl group (-H) at the 3′ carbon. |
| 2) Use in Sequencing | Standard nucleotide used in regular DNA replication. | Used in Sanger sequencing to terminate DNA chain elongation at specific points |
| 3) Nucleotide Components | Contains a phosphate group, a deoxyribose sugar, and a nitrogenous base (A, T, C, or G). | Similar to deoxyribonucleotide but lacks the 3′ hydroxyl group on the sugar. |
| 4) Role in DNA Synthesis: | Used in the synthesis of DNA by forming phosphodiester bonds. | OH group inhibits the addition of subsequent nucleotides. |
b) When performing a western blot, what is the purpose of adding a secondary antibody?
Ans→ To bind to the primary antibody, amplifying the signal and allowing for the detection of the target protein by linking it to a detectable marker, such as an enzyme or fluorophore.
c) How photo dimers in DNA is repaired?
Ans→ Photodimers in DNA, such as thymine dimers caused by UV light, are repaired by nucleotide excision repair (NER). In this process, the damaged DNA segment is removed and replaced with correct nucleotides using the undamaged strand as a template.
d) Which processes in protein synthesis require hydrolysis of GTP? What is Shine-Dalgarno sequence?
Ans→ In the Elongation process of protein synthesis, GTP hydrolyzes rapidly on EF-Tu.
The Shine-Dalgarno sequence is a short, conserved purine-rich sequence in prokaryotic mRNA, typically located just upstream of the start codon.
e) Which bond is used to stabilize the double helix of DNA? What is Tm?
Ans→ Hydrogen bond i used to stabilize the double helix of DNA.
f) Why do you think that most promoter regions are A-T rich?
Ans→ Most promoter regions are A-T rich because A-T base pairs have only two hydrogen bonds, making them easier to separate compared to G-C pairs. This allows easier access for the transcription machinery to bind and initiate transcription.
g) What are the differences between Group I and Group II introns?
Ans→
| Group I Introns | Group II Introns | |
| Splicing Mechanism | Self-splicing via a guanosine cofactor (no lariat formation). | Self-splicing with lariat intermediate formation. |
| Structure | Less complex secondary structure | More complex secondary structure with domains aiding catalysis. |
| Splice Sites | Cleavage occurs with the help of an external guanosine nucleotide. | Uses internal adenosine as a branch point for lariat formation. |
| rRNA genes in Tetrahymena. | Introns in fungal mitochondrial genes. |
h) What do you mean by ‘coupled transcription-translation’.
Ans→ Coupled transcription-translation refers to the simultaneous process of transcription and translation in prokaryotes, where as soon as mRNA is synthesized, ribosomes begin translating it into protein, without a significant delay.
Molecular Biology 2022
1.) a) Mention the role of “Elongation factor thermo unstable”.
Ans→ EF-Tu or Elongation factor thermal unstable catalyzes the binding of amino acyl tRNA to the A site of the ribosome during elongation process of protein synthesis.
b) Name one staining agent of RNA.
Ans→ Toluidine blue
c) Mention the significance of CCA tail in tRNA.
Ans→ CCA tail at 3’ end of the tRNA provide the binding site of a specific amino acid by the amino-acyl tRNA synthetase enzyme.
d) Mention the significance of XPC enzyme.
Ans→ (i) XPC Enzyme function in DNA damage recognition
(ii) XPC proteins is recruited to eliminate the DNA damage.
e) Draw the difference between Thymine and Uracil.
Ans→
f) Write down the repeated sequence in human DNA, that helps telomere replication.
Ans→ CCCTAA/TTAGGG
g) Where you find the 5′ to 5′ triphosphate linkage?
Ans→ At the 5’ end of mRNA bind with 5’ of Guanine nucleotide.
h) What is Tm?
Ans→ The temperature at which one half of DNA duplex will dissociates to become single stranded DNA and duplex stability called melting temperature or Tm
i) What is replisome?
Ans→ A replisome is a complex of proteins involved in DNA replication, responsible for unwinding the DNA and synthesizing new strands during cell division.
j) What is riboswitch?
Ans→ Riboswitches are structured non-coding RNA domains that selectively bind metabolites and control gene expression.
k) What is antisense RNA?
Ans→ Antisense RNA is a single stranded RNA formed by using ‘sense strand’ of DNA that is complementary to protein coding mRNA.
l) What type of PCR is most efficient for the detection and quantification of mRNA expression?
Ans→ Real-Time PCR
m) Do the Restriction endonucleases serve as primitive form of immune system in bacteria?
Ans→ Yes, Restriction endonuclease as primitive form of immune system in bacteria. This enzymes play a crucial role in preventing viral infections especially from bacteriophages by cleaving the foreign DNA.
n) What are the most conserved and least conserved Histones?
Ans→ Conserved Histone: Histone H4
List conserved Histone: Histone H1
o) What do you understand by ‘Guide RNA’?
Ans→ Guide RNA (gRNA)is a type of RNA molecule helps in guiding of RNA-DNA target enzymes.
- Answer any five of the following questions:
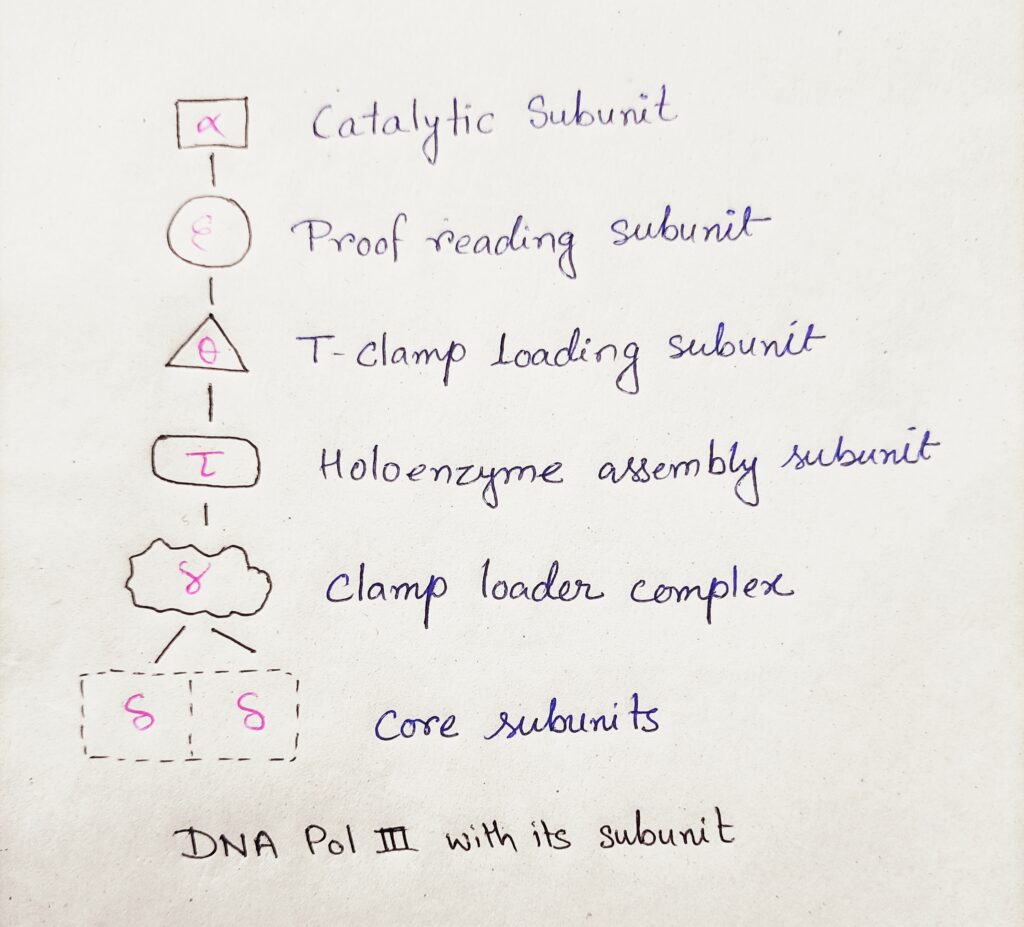
a) Draw a line diagram of DNA Pol-Attenuation III enzyme of E.coli with its subunits.
Ans→
b) Explain the annealing step in PCR.
Ans→ The annealing step in PCR is when the temperature is lowered to allow the primers to bind (anneal) to their complementary sequences on the single-stranded DNA template. This step typically occurs at 50-65°C.
c) efficiency is correlated with the stability of secondary structure embedded in TrpL. – Justify the statement.
Ans→ The efficiency of transcription in the TrpL (tryptophan leader) region is influenced by the stability of its secondary structures, as these structures control the formation of terminator or antiterminator elements. Stable secondary structures can promote termination of transcription, while less stable structures allow transcription to proceed, impacting gene expression efficiency.
d) Define Ori C.
Ans→ OriC is the origin of replication in bacteria, specifically in Escherichia coli, where DNA replication begins. It is a specific sequence of DNA that signals the start of the replication process.
e) What is the fate of DNA in acidic solution?
Ans→ In an acidic solution, DNA tends to lose its negative charge as the phosphate groups on the backbone become protonated, which can lead to the destabilization of its double helix structure and cause the strands to separate.
f) If a DNA segment contains 200 nucleotide pairs then what is the length of the DNA segment and calculate the number of spirals in the DNA.
Ans→
g) What is SOS repair?
Ans→ SOS repair is a quick, emergency solution to fix a critical issue, often in a technical or mechanical context, to restore basic functionality until a more permanent fix can be applied.
h) Mention the role of ‘hydrolysis of GTP’ in protein synthesis.
Ans→ The hydrolysis of GTP in protein synthesis provides energy for several key steps, including the binding of tRNA to the ribosome, the elongation of the polypeptide chain, and the movement of the ribosome along the mRNA. It ensures the accuracy and efficiency of translation by driving the conformational changes required for these processes.
